With a focus on correcting the non-Asian bias in genetic research, a paper in the multidisciplinary scientific publication, Nature, will have implications pertaining to our understanding of the formation of the population of Asia, and on medicine and healthcare in the region.
The article also throws up interesting facets about how the urban population of Chennai has largely remained isolated and how the Indo-European language speakers from Pakistan have the least in common with Denisovans, an earlier ancestor.
The article is a result of the largest genomic study conducted among 1,739 Asian individuals from 219 populations across the continent. The 598 individuals sampled from India was the highest from a single country. The study has been undertaken by the US and Asian scientists from Nanyang Technological University, Singapore, National Institute of Biomedical Genomics (NIBG), Kalyani, India, the University of California in the USA with the aid of private players from the bio-technology industry. The group calls itself the GenomeAsia 100K consortium and the article is on the cover of Nature this week.
Advertisement

The country-wise breakdown of the number of DNA samples taken
According to Partha P. Majumder, Founder of NIBG, Kalyani, and one of the co-authors of the paper, the reason why an extensive study of Asian DNA is necessary is due to an absence of data about the Asian genomes. “It has been the experience that many points embedded on currently available DNA chips are non-informative for Asian individuals. In India, about 30-40% of these points are non-informative. This implies that use of DNA-chips optimised for the Caucasian populations is quite wasteful in the Asian context,” he writes in a statement shared with Outlook.
Advertisement
Majumder explains that the objectives of the study were three-fold:
- to generate and catalogue DNA sequencing and variation on a large population of Asian individuals
- whether insights can be drawn from whole-genome sequence of databases
- to draw relevant medical conclusions from the data
The scientists have found “200,000 previously unreported novel DNA variants among Asians” and that “23% of protein-altering variants found in Asia are unreported in existing databases.” Majumder explains that the data is important in order to discover genes associated with diseases common in Asia. Proteins are also important since changes in protein are related to diseases.
Using the genetic data at their disposal the scientists were able to reveal a DNA variant in a gene (NEUROD1) which possibly responsible for a particular kind of diabetes. Another DNA variant in the haemoglobin gene has been linked to beta-thalassemia found only in South Indians, says Majumder. He adds that the most notable finding of the study is that Carbamazepine, an anti-convulsant used to treat medical disorders, may have adverse effects on about 400 million South-East Asians who form part of the Austronesian language group.
The scientists have also addressed how populations were formed in Asia. About 4 million years ago, two precursors to modern-day humans, the Neanderthal and Denisovan came about. The Homo Sapien that came about around 1.5 million years ago, today contains high admixture with the other two, the degree of mixing varying across Asia. Majumder says that in India, the researchers found that Tribal and non-Indo European speakers had more Denisovan DNA than the non-tribals and Indo-European speakers. He further says that upper-caste groups have lesser Denisovan DNA and that Indo-European speakers of Pakistan had the lowest Denisovan component.
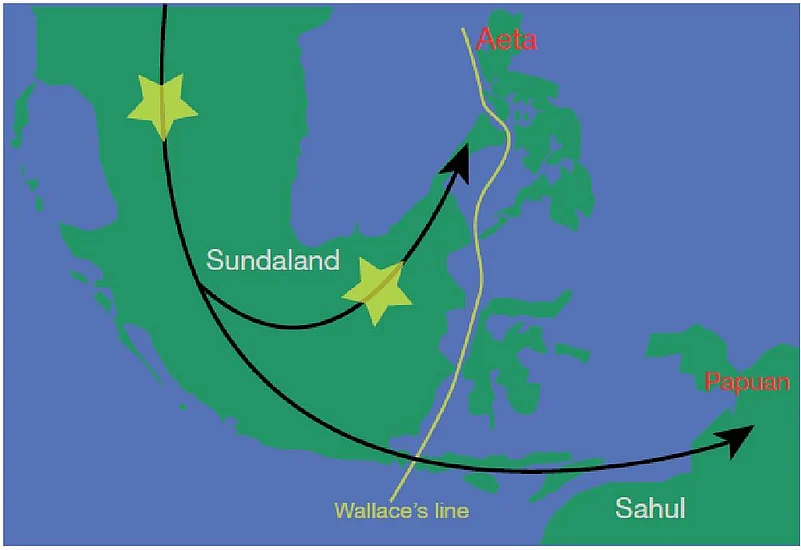
Proposed modern human migration route into Southeast Asia about 20,000 years ago with potential locations of admixture of modern humans with the archaic human known as Denisovan (yellow asterisks). Green indicates the above water landmass at the glacial maximum and white outlines indicate present-day shorelines
Advertisement
“The simplest explanation of these findings is that Indo-European speaking migrants who entered the Indian subcontinent from the northwest admixed with an indigenous group ancestral to present-day south Indians; the ancestral group had a high level of Denisovan admixture,” he writes.
With regard to genetic structures of the Asian population, the research says that the current people are a result of admixtures with 14 ancestral populations in the past with “five predominant ancestral admixture events seem to have taken place to give rise to the present-day Indian populations, including the populations of Andaman & Nicobar islands.”
The researchers also discovered that some large tracts of urban India, like Chennai, show characteristics of genetically isolated populations. “It is, therefore, possible that some urban populations may have arisen from a small number of founders and then numerically expanded quickly,” Majumder explains.